Introduction
Fusion genes (FGs) are major molecular biological abnormalities in acute leukemia and have been used as molecular markers for the diagnosis, classification, risk stratification and targeted therapy of leukemia. We previously reported common FGs were presented in approximately 41% of acute myeloid leukemia (AML) cases (Chen X et al., Leuk Lymphoma 2019). The rapid development of sequencing technology and the decline of sequencing costs in recent years have made whole transcriptome sequencing (WTS) more accessible, which can not only analyze known FGs, but also has unique advantages in identifying unknown rare and variant FGs. We aimed to identify novel fusion transcripts with clinical relevance and delineate a comprehensive map of FGs in AML based on a large cohort using WTS.
Methods
We studied 400 consecutively diagnosed AML patients using WTS, 50 normal bone marrow (BM) samples from healthy donors were used as controls. Written informed consents were obtained from all patients and healthy donors or their guardians in accordance with the Declaration of Helsinki.
WTS was performed using RNA extracted from the BM samples by HiSeq 2500. Reads were mapped and processed by Arriba (v1.0.1) to generate gene fusions. Only in-frame fusions of high confidence were retained. We applied FGs to a four-tier system as follows; (A) pathogenic: well-known FGs or new members of common fusion gene families (FG-FMs) with definite pathogenicity in hematological malignancies or other tumors. (B) likely pathogenic: rarely reported FGs or new members of rare FG-FMs in hematological malignancies or other tumors without functional verification. (C) uncertain significance: novel FGs and both genes have not been reported in tumors. (D) non-pathogenic: FGs detected in normal samples.
Results
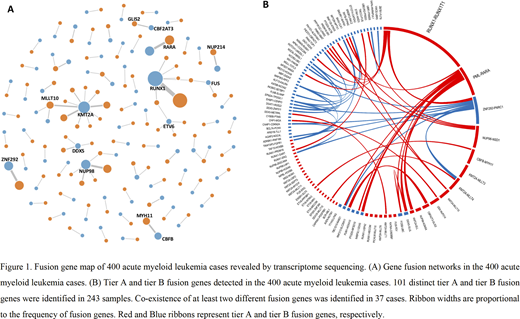
Our analysis identified 342 high confidence in-frame FGs in 400 AML cases. We further classified the FGs into four ties based on pathogenicity and the 199 tier A and 87 tier B FGs were adopted to the final FG list for further analysis (Figure 1A).
The 286 tier A and tier B FGs were identified in 243 (61%) samples (mean, 1.2 per sample), of which 101 were distinct events. Tier A FGs were detected in 197 (49%) cases while 46 (12%) cases had tier B FGs without tier A FGs. The remaining 157 (39%) cases had no tier A nor tier B FGs. We identified 37 cases with co-existence of at least two different FGs, accounting for 9% of all cases enrolled in this study and 15% of all positive cases (Figure 1B). Multiplex-nested RT-PCR which was designed to detect 41 common FGs (all belonged to tier A FGs) was performed in all 400 cases and only 166 (42%) cases were positive.
We found 27 kinds of recurrent FGs which occurred at least twice, including 18 tier A and 9 tier B FGs, respectively. Of these, 7 recurrent FGs have never been previously reported. Furthermore, we classified the 101 distinct FGs found in the 243 cases according to FG-FMs, which referred to FGs that involve one protagonist gene and multiple fusion partners. Nearly half (47%) FGs could be classified into 18 FG-FMs, such as RUNX1-FM, KMT2A-FM, NUP98-FM, RARA-FM, ZNF292-FM, DDX5-FM, and NUP214-FM. The other 54 distinct FGs like CBFB-MYH11, CBFA2T3-GLIS2, and KAT6A-CREBBP could not be classified into any family. Most FGs which could not be clustered into FG-FMs occurred only once. All in all, 74% of the 286 tier A and tier B FGs could be classified into FG-FMs, the remaining 26% FGs mainly belonged to tier B and rarely recurred in different samples. When we focused on tier A FGs, 90% of them could be clustered into FG-FMs, while only 10% of them could not be classified into any FG-FM.
Conclusions
We described the map of FGs detected in a large cohort of AML and revealed FGs with clinical relevance that have not been previously recognized. Classifying FGs according to FG-FMs can better understand their pathological significance and suggest new classification patterns of acute leukemia. WTS is a valuable tool and should be widely used in the routine diagnostic workup of AML.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal